Olympic gels formed through catenation of dsDNA rings regulated by topoisomerase II: A coarse-grained model
 featured.jpg
featured.jpg
Abstract
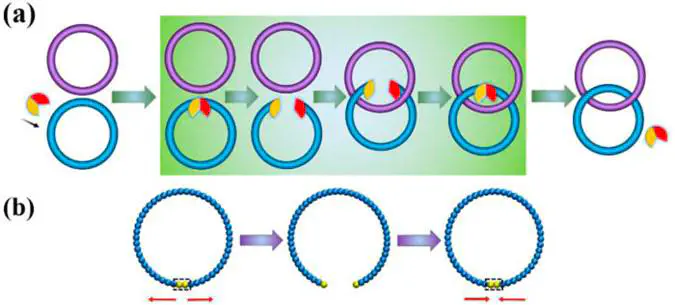
Topological regulation of DNA by topoisomerases in cells is very crucial for life. We propose a coarse-grained model to study the catenation process of double-stranded DNA (dsDNA) rings regulated by topoisomerase II (TOP2) and provide a computational method to characterize the topological structures of the Olympic gels obtained. The function of TOP2 in the catenation of dsDNA rings is implicitly fulfilled by operating the length of a stretchable catch bond in the dsDNA ring. After the catenation reaction of initially noncatenated dsDNA rings in the solution, the Olympic gel is obtained and the interlocked topology of the dsDNA rings can be characterized by a computational method derived from the HOMFLY polynomial, based on which the catenation degree and the complexity of catenation are quantified. Detailed dependence of the catenation degree and the complexity of the catenated topology on key parameters, including the size of the transient broken gap and the duration time of the break on the dsDNA ring during operation by TOP2, the initial molar ratio of TOP2 to the dsDNA rings, and the reaction temperature, has been investigated.